 Daily Newsletter
Daily Newsletter
November 6, 2012 Lac Operon
One of the most well studied gene regulation systems is the Lac Operon found in Escherichia coli. To understand this system, it is important to first understand that bacteria do not experience Transcription and Translation in exactly the same way as eukarotes (the mechanics are very similar, but there are some distinct differences).
The core difference between prokaryotes and eukaryotes is the nucleus. Since bacteria lack a nucleus, transcription takes place in the cytosol. There is no need for a tag to get out of the nucleus, so there is no need for capping. Also, there are no introns, so no need for splicing. Basically, there is no processing of RNA to make mRNA. There is no pre-mRNA. When a gene is transcribed, the transcript is mRNA.

Since there is no membrane separating transcription from translation, you can couple these to processes. As mRNA is made, it can be translated (polyribosome).
Bacteria have a single circular molecule of DNA (a genophore, not a chromosome). They have to conserve their genetic space, so bacteria combine genes for a metabolic process in a single mRNA. IMPORTANT: bacteria can combine genes for a metabolic pathway into a sequential sequence with a single promoter. Thus, when you transcribe, you get all the genes for a given metabolic pathway.
The word OPERON describes this unique arrangement of prokaryotic genes: One promoter and one operator for a given series of metabolically linked genes.

The lac Operon holds three genes that give the cell the ability to take in and use the sugar LACTOSE. [Image Note: Bacteria have two promoter regions, -10 & -35, for a given gene or operon]
For E. coli, glucose is the preferred sugar. When glucose is present, there is no need to use lactose: these genes are not transcribed. When there is no glucose, E. coli has to use other sugars. IF lactose is present, the genes for lactose utilization will be made. Conversely, if there is no lactose, they genes remain locked down.
- Glucose Present: No transcription
- Glucose Absent: Minimal transcription
- Glucose Absent, Lactose Present: Transcription of the lac operon.
- You can block the operator of the gene. This prevents RNA polymerase from making RNA.
- You can alter the promoter (or the interaction between transcription factors and DNA) to prevent binding of the Transcription Complex (RNA polymerase).
As you can see in the image to the right, the lac repressor is a protein that is constitutively (always) expressed. This indicates that the lac operon is normally turned OFF. Notice that the gene for the regulatory protein is upstream from the lac operon.
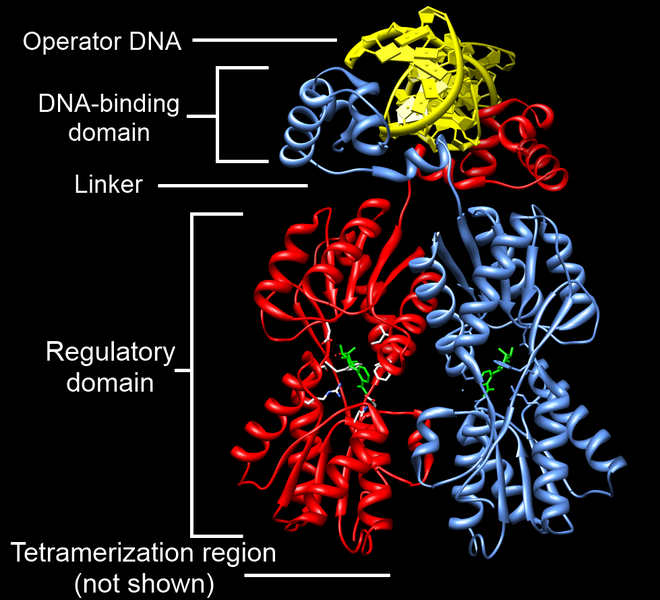 The lac repressor (LacI) binds to the major groove of the DNA at the operator. As you can see in the annotated image of the repressor, you have a DNA-binding region (active site), and a regulatory domain. The regulatory domain has the ability to bind to Lactose (the inducer).
The lac repressor (LacI) binds to the major groove of the DNA at the operator. As you can see in the annotated image of the repressor, you have a DNA-binding region (active site), and a regulatory domain. The regulatory domain has the ability to bind to Lactose (the inducer).You must have a way to unlock the operon, or to put it another way, to inactivate the repressor. An inducer is a ligand that can bind to a regulatory domain, changing the shape of the regulatory domain, and thus inactivating the DNA-binding domain. In the case of the the lac repressor, the ligand is allolactose (a derivative of lactose). When allolactose binds to the lac repressor, the repressor is inactivated, and the operon cleared.
Therefore, the lac operon is partially regulated by the presence of lactose in the environment. If there is no lactose in the environment, then there is no need to transcribe the three genes needed to use lactose.
Remember, we don't want to expend energy for things we don't need. Below is the size of the three gene products:
- β-Galactosidase 1,024 Amino Acids
- β-Galactoside Permease 418 Amino Acids
- β-galactoside transacetylase 203 Amino Acids
Tomorrow we will look at the positive regulation of the lac operon. Remember, you still will not transcribe this operon if glucose is present. You only transcribe when glucose is absent. So how does the operon know when glucose is absent? That will be our discussion tomorrow.
Daily Challenge
In your own words, describe how lactose (allolactose) is used to regulate the transcription of the lac operon. In your discussion, make sure that you explain the concept of an operon, and discuss the differences between eukaryote and prokaryote gene transcription.Link to Forum




 In eukaryotic DNA every gene starts with a promoter. This is a sight of ~8 nucleotides visible in the major groove of DNA. The transcription complex recognizes this sequence as a "START" indicator. The main core of the transcription complex will be RNA polymerase. This enzyme works to build a strand of RNA complementary to DNA.
In eukaryotic DNA every gene starts with a promoter. This is a sight of ~8 nucleotides visible in the major groove of DNA. The transcription complex recognizes this sequence as a "START" indicator. The main core of the transcription complex will be RNA polymerase. This enzyme works to build a strand of RNA complementary to DNA. 






 molecule. As can be seen in the image to the right, the major groove is wide enough to "see" the base pairs. The base pairs have an electrochemical profile, and thus can respond to other chemicals (via van Der Waals forces). Thus,
molecule. As can be seen in the image to the right, the major groove is wide enough to "see" the base pairs. The base pairs have an electrochemical profile, and thus can respond to other chemicals (via van Der Waals forces). Thus, 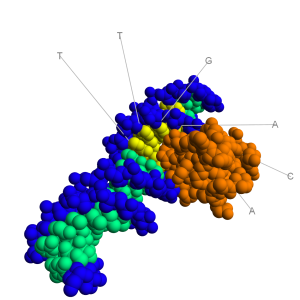 The image to the right shows a bacterial promoter event. One of the factors needed to start transcription (by recognizing the promoter) has bound into the major groove. This recognition event is needed to identify the start point of a gene. The Transcription Initation Complex will then begin to form at this site, and begin the transcription of the gene.
The image to the right shows a bacterial promoter event. One of the factors needed to start transcription (by recognizing the promoter) has bound into the major groove. This recognition event is needed to identify the start point of a gene. The Transcription Initation Complex will then begin to form at this site, and begin the transcription of the gene.  House keeping genes are those that are needed for the general function of the cell, and can include genes for glycolysis, citric acid cycle, and ribosomes. These genes are always ON, and are referred to as
House keeping genes are those that are needed for the general function of the cell, and can include genes for glycolysis, citric acid cycle, and ribosomes. These genes are always ON, and are referred to as 

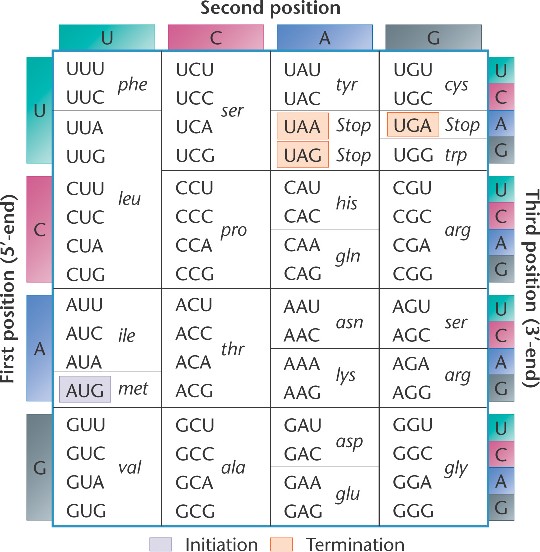

 DNA is a double stranded molecule, in which the strands are anti-parallel. This means that one strand starts at the 5' end
DNA is a double stranded molecule, in which the strands are anti-parallel. This means that one strand starts at the 5' end 
 At this point, the critical thing to remember is that DNA Polymerase will need a free 3' end on which to add a new nucleotide.
At this point, the critical thing to remember is that DNA Polymerase will need a free 3' end on which to add a new nucleotide.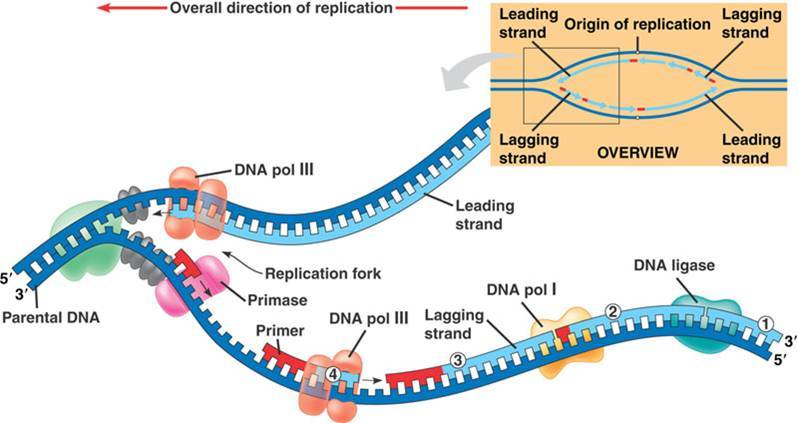 The image above shows the leading and lagging strand. Notice that the leading strand is replicating toward the
The image above shows the leading and lagging strand. Notice that the leading strand is replicating toward the  one from their mother and one from their father. These two individual examples of chromosome 1 are considered homologous. Homologous chromosomes carry the same genes, but each homolog (one copy) will have a unique set of alleles (one allele for each gene).
one from their mother and one from their father. These two individual examples of chromosome 1 are considered homologous. Homologous chromosomes carry the same genes, but each homolog (one copy) will have a unique set of alleles (one allele for each gene).  IMPORTANT NOTE: Remember that what we are seeing are chomosomes. The two chromatids of a given chromosome are the products of DNA replication. The image to the left should help you remember that you start with a Maternal DNA molecule and a Paternal DNA molecule. After DNA replication, these will consense into maternal and paternal chrmosomes. It is critical that you remember the difference between Chromosome and Chromatid.
IMPORTANT NOTE: Remember that what we are seeing are chomosomes. The two chromatids of a given chromosome are the products of DNA replication. The image to the left should help you remember that you start with a Maternal DNA molecule and a Paternal DNA molecule. After DNA replication, these will consense into maternal and paternal chrmosomes. It is critical that you remember the difference between Chromosome and Chromatid. Prophase 1 (prophase of Meiosis I) is the most critical stage of meiosis. Prior to the reduction division (which occurs in Anaphase 1), a
Prophase 1 (prophase of Meiosis I) is the most critical stage of meiosis. Prior to the reduction division (which occurs in Anaphase 1), a 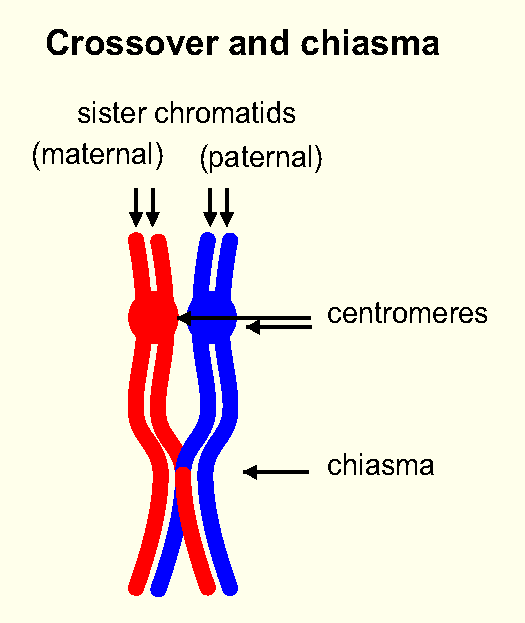 This recombination event begins when homologous chromosomes are brought together during prophase/prometaphase I. The homologs possess DNA sequence similarities, and are able to bind to each other (the homologs chemically recognize each other). Regions where Cross-Over (recombination) can occur begin to over lap, forming a visible structure known as a
This recombination event begins when homologous chromosomes are brought together during prophase/prometaphase I. The homologs possess DNA sequence similarities, and are able to bind to each other (the homologs chemically recognize each other). Regions where Cross-Over (recombination) can occur begin to over lap, forming a visible structure known as a 

 NOTICE: each cell produced in meiosis I now undergo meiosis II.
NOTICE: each cell produced in meiosis I now undergo meiosis II.  Daily Newsletter
Daily Newsletter Interphase is divided into three distinct steps: G1 Phase, S Phase, and G2 Phase.
Interphase is divided into three distinct steps: G1 Phase, S Phase, and G2 Phase.
 Throughout the cell cycle, there are time points referred to as
Throughout the cell cycle, there are time points referred to as