Special Newsletter
Gibbs Free Energy and Transition State Energy
This goal of this newsletter is to help students navigate the complexity of Gibbs Free Energy (ΔG) and help avoid confusion about Gibbs Free Energy and Transition State Energy (Energy of Activation).
Gibbs Free Energy (ΔG) is a thermodynamics concept, and describes the thermodynamic potential of a system. It measures the "work" that can be obtained from a system, for us a chemical reaction, if that system is held at constant temperature and pressure. The goal is to determine whether a given reaction is spontaneous in the direction written, i.e., whether the reaction will proceed in the given direction without the input of energy.
Mathematically, Gibbs Free Energy is described as:
ΔG=ΔH−TΔS
Where:
- ΔG The change in Free Energy
- ΔH The change in Enthalpy
- ΔS The change in Entropy
Gibbs free energy is a measure ONLY of the difference in free energy of the products and reactants, and does not tell us about the rate of the reaction. It only tells us whether the reaction is Exergonic or Endergonic.
- ΔG < 0 Exergonic: The reaction is considered spontaneous in the direction written.
- ΔG = 0 The system is in equilibrium
- ΔG >0 Endergonic: To carry out this reaction as written, there needs to be an addition of free energy (NOTE: this means that the reverse reaction is spontaneous).
Transition State Energy is not Gibbs Free Energy.
Gibbs Free Energy does not describe the path of transformation or the mechanism of transformation. It does not describe the rate of transformation. It only describes whether the reaction is spontaneous, non-spontaneous or at equilibrium.
The breakdown of the dissacharide sucrose to glucose and fructose has a ΔG of -5.5 kcal/mol. This is a spontaneous (Exergonic) reaction in terms of ΔG. Yet you can store sucrose in your kitchen and it remains sucrose. There is no spontaneous degradation into monosaccharides. Why?
Sucrose is a stable molecule. In order to force the breakdown (catabolism) of sucrose, we need to destabilize the molecule. This destabilization is the transition state of the reaction, and in a closed system, requires the input of energy. This is termed the Activation Energy of the reaction (you will also hear this described as the Transition State Energy). If we were to look at our breakdown of sucrose, we would see the following:
Sucrose ⇄ Transition State → Glucose + Fructose
The energy of the transition state is noted as either Ea or ΔG‡. These two expression are from different formula for calculating activation energy. ΔG‡ is used in a formula that relates activation energy to Gibbs Free Energy (the Eyring equation). In either case, the expression describe transition state energy (aka, activation energy).
In biochemistry, enzymes are used to reduce the activation energy (just like catalysts are used in chemistry). Catalysts and Enzymes reduce the activation energy ΔG‡; they do not alter Gibbs Free Energy (ΔG). This is a critical concept!
Transition state deals with the rate of the reaction. By lowering the activation energy (transition state energy), you increase the rate of the reaction. In essence, you are making the reaction more likely to happen. But the enzyme does not change Gibbs Free Energy.
 |
| Original Image from ChemWiki, http://chemwiki.ucdavis.edu/@api/deki/files/10017/fREE_eNERgY_cHART.jpg?size=bestfit&width=471&height=294&revision=1 |
Take home message:
Gibbs Free Energy (ΔG) describes whether a reaction is exergonic or endergonic. It does not describe rate, and neither enzymes or catalysts will alter ΔG.Activation Energy (ΔG‡) does not alter ΔG; it does not determine whether a reaction is spontaneous or non-spontaneous. Activation energy does help determine the rate of the reaction.

 Daily Newsletter
Daily Newsletter

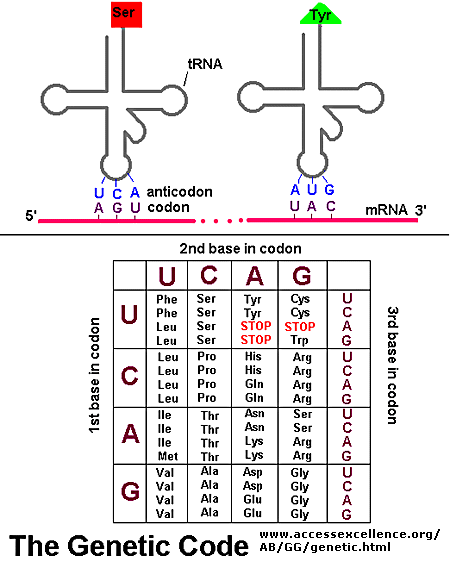




 molecule. As can be seen in the image to the right, the major groove is wide enough to "see" the base pairs. The base pairs have an electrochemical profile, and thus can respond to other chemicals (via van Der Waals forces). Thus,
molecule. As can be seen in the image to the right, the major groove is wide enough to "see" the base pairs. The base pairs have an electrochemical profile, and thus can respond to other chemicals (via van Der Waals forces). Thus,  The image to the right shows a bacterial promoter event. One of the factors needed to start transcription (by recognizing the promoter) has bound into the major groove. This recognition event is needed to identify the start point of a gene. The Transcription Initation Complex will then begin to form at this site, and begin the transcription of the gene.
The image to the right shows a bacterial promoter event. One of the factors needed to start transcription (by recognizing the promoter) has bound into the major groove. This recognition event is needed to identify the start point of a gene. The Transcription Initation Complex will then begin to form at this site, and begin the transcription of the gene.  House keeping genes are those that are needed for the general function of the cell, and can include genes for glycolysis, citric acid cycle, and ribosomes. These genes are always ON, and are referred to as
House keeping genes are those that are needed for the general function of the cell, and can include genes for glycolysis, citric acid cycle, and ribosomes. These genes are always ON, and are referred to as 

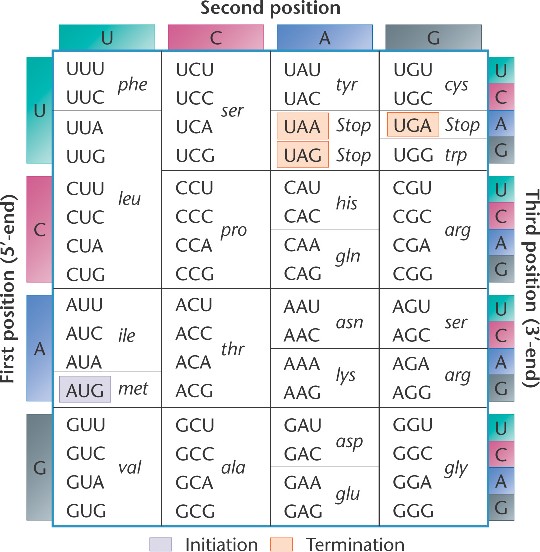
 DNA is a double stranded molecule, in which the strands are anti-parallel. This means that one strand starts at the 5' end
DNA is a double stranded molecule, in which the strands are anti-parallel. This means that one strand starts at the 5' end 
 At this point, the critical thing to remember is that DNA Polymerase will need a free 3' end on which to add a new nucleotide.
At this point, the critical thing to remember is that DNA Polymerase will need a free 3' end on which to add a new nucleotide.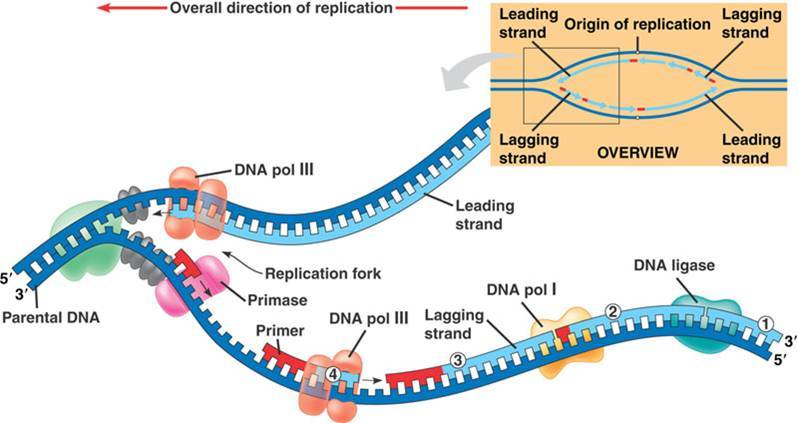 The image above shows the leading and lagging strand. Notice that the leading strand is replicating toward the
The image above shows the leading and lagging strand. Notice that the leading strand is replicating toward the 



 Glycerol is a 3 carbon compound that we will see from time to time. It acts as the backbone or schaffold of the triglycerides and phospholipids. As you can see, on each carbon atom, there is a hydroxyl group (-OH). This hydroxyl group is where other molecules can bond. Another thing to note is that there is free rotation around the carbon atoms. When you take organic chemistry, you will learn more about free rotation, why it is important, and how it can affect a molecule. For now, just note that there can be free rotation.
Glycerol is a 3 carbon compound that we will see from time to time. It acts as the backbone or schaffold of the triglycerides and phospholipids. As you can see, on each carbon atom, there is a hydroxyl group (-OH). This hydroxyl group is where other molecules can bond. Another thing to note is that there is free rotation around the carbon atoms. When you take organic chemistry, you will learn more about free rotation, why it is important, and how it can affect a molecule. For now, just note that there can be free rotation. chemicals are depicted. Below the first depiction is a molecular model. Carbon atoms are in black, and the white balls are hydrogens. What you will begin to recognize in these diagrams is that there are only single bonds between the carbon atoms. This means that the maximum number of hydrogen atoms are attached to the fatty acid. In other words, they are saturated with hydrogen.
chemicals are depicted. Below the first depiction is a molecular model. Carbon atoms are in black, and the white balls are hydrogens. What you will begin to recognize in these diagrams is that there are only single bonds between the carbon atoms. This means that the maximum number of hydrogen atoms are attached to the fatty acid. In other words, they are saturated with hydrogen. Note that there is a double bond in the carbon chain. Notice that the chain is bent, or kinked. This creates a very different structure for lipids that carry unsaturated fatty acids.
Note that there is a double bond in the carbon chain. Notice that the chain is bent, or kinked. This creates a very different structure for lipids that carry unsaturated fatty acids. As you can see in the diagram, the two molecules are joined together through an oxygen molecule. As with other biosynthetic reactions, this is a dehydration synthesis (water is released). The resulting bond, as noted in the diagram, is an
As you can see in the diagram, the two molecules are joined together through an oxygen molecule. As with other biosynthetic reactions, this is a dehydration synthesis (water is released). The resulting bond, as noted in the diagram, is an  Recall the look of the glycerol, and note that there are three locations where an ester bond can be formed. In a triglyceride, a fatty acid will be bound to each of the carbon atoms by ester bonds. Tri- means three, so we have three fatty acids attached to the glycerol. The image to the right is an example of a triglyceride, and please note, you can have more than one type of fatty acid in a triglyceride.
Recall the look of the glycerol, and note that there are three locations where an ester bond can be formed. In a triglyceride, a fatty acid will be bound to each of the carbon atoms by ester bonds. Tri- means three, so we have three fatty acids attached to the glycerol. The image to the right is an example of a triglyceride, and please note, you can have more than one type of fatty acid in a triglyceride. Phosphytidyl choline is a commonly studied phospholipid that uses choline as the charged organic structure. NOTE: both the phosphate group and the organic structure carry a charge. This phosphate head is
Phosphytidyl choline is a commonly studied phospholipid that uses choline as the charged organic structure. NOTE: both the phosphate group and the organic structure carry a charge. This phosphate head is 
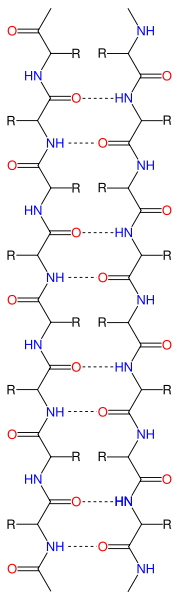 Notice that in the image there are yellow dashed lines. These represent hydrogen bonds (van der Waals forces) between amino acids. The green ribbon represents the backbone of the amino acid chain (amino-chiral-carboxyl connected to amino-chiral carboxyl and so on). So the interactions (notably from polar partially charged side chains has produced a twist in the primary structure.
Notice that in the image there are yellow dashed lines. These represent hydrogen bonds (van der Waals forces) between amino acids. The green ribbon represents the backbone of the amino acid chain (amino-chiral-carboxyl connected to amino-chiral carboxyl and so on). So the interactions (notably from polar partially charged side chains has produced a twist in the primary structure.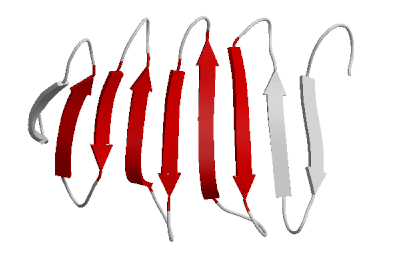 In this diagram, the red arrows represent a portion of the primary structure that has begun to form β-pleates. Notice that six red arrows are arranged together. The purpose of this diagram is to show the placement of the β-pleated sheet. Consider a sheet. It is flat with two sides. Why do you think it would be important for a protein to fold in such a way as to create a relatively "flat" surface with two sides?
In this diagram, the red arrows represent a portion of the primary structure that has begun to form β-pleates. Notice that six red arrows are arranged together. The purpose of this diagram is to show the placement of the β-pleated sheet. Consider a sheet. It is flat with two sides. Why do you think it would be important for a protein to fold in such a way as to create a relatively "flat" surface with two sides?
 The amino acid cystine contains a thiol (-SH). The thiols of two cystines can react to form a
The amino acid cystine contains a thiol (-SH). The thiols of two cystines can react to form a  (like collagen). There is a final level of structure. Some functional proteins are actually made up of multiple individual proteins. A great example of this is hemoglobin. The hemoglobin molecule, seen to the left, is composed of four individual proteins: 2 α-hemoglobin (red) and 2 β-hemoglobin (blue). The green structure is Heme, a prosthetic group that is used to hold oxygen, and is attached by electrostatic interactions (van der Waals forces) to the proteins.
(like collagen). There is a final level of structure. Some functional proteins are actually made up of multiple individual proteins. A great example of this is hemoglobin. The hemoglobin molecule, seen to the left, is composed of four individual proteins: 2 α-hemoglobin (red) and 2 β-hemoglobin (blue). The green structure is Heme, a prosthetic group that is used to hold oxygen, and is attached by electrostatic interactions (van der Waals forces) to the proteins.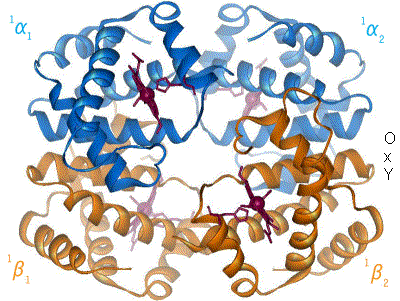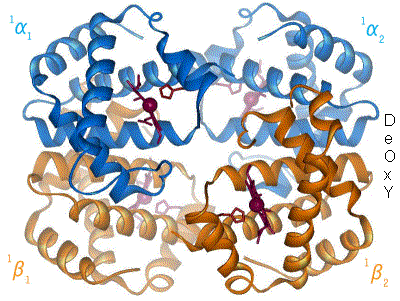



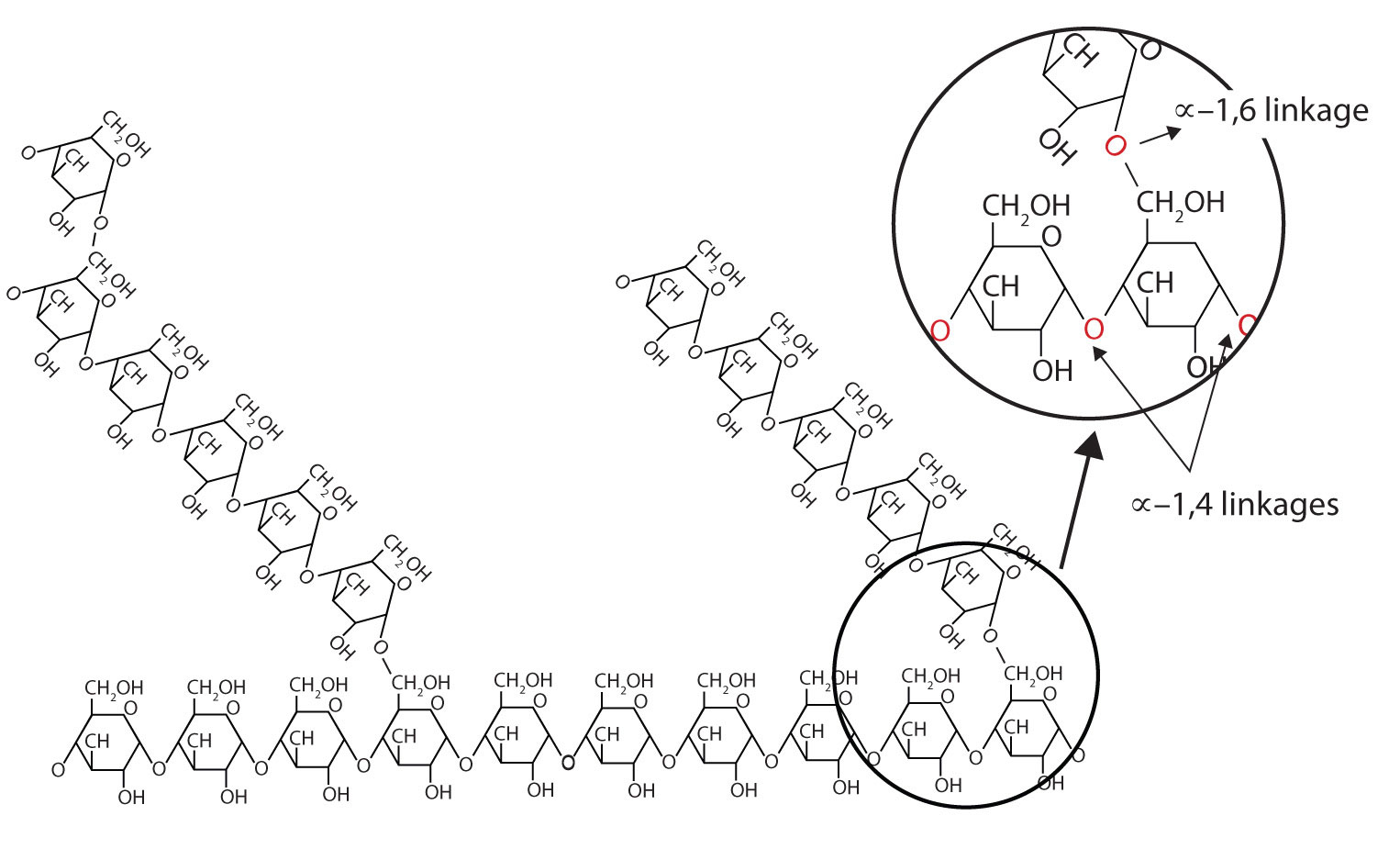


 The base molecule is needed to link amino acids together into a polymer. A condensation (dehydration synthesis) reaction is used to form peptide bonds, the specific bond type that links amino acids together. [NOTE: biopolymers (save for lipids which are not polymers) have specific names for the bonds between monomers.] In the formation of a peptide linkage (bond), you will have a carboxyl and amino group linking together, with water being a product.
The base molecule is needed to link amino acids together into a polymer. A condensation (dehydration synthesis) reaction is used to form peptide bonds, the specific bond type that links amino acids together. [NOTE: biopolymers (save for lipids which are not polymers) have specific names for the bonds between monomers.] In the formation of a peptide linkage (bond), you will have a carboxyl and amino group linking together, with water being a product.