 Microbiology Daily Newsletter
Microbiology Daily Newsletter
February 20, 2014 - Transcription Termination
Transcription Termination
The elongation phase of transcription (polymerization of RNA) occurs the same in all domains, working via base complementarity. Termination, like initiation, is different. In bacteria, transcription termination occurs either through an intrinsic termination system (Rho-Independent) or through the use of a Rho (ρ) factor (Rho-Dependent).The Intrinsic System (Rho-Independent)
After a the coding region of the gene, or the final coding region of an operon, there will be an inverted repeat of nucleotides on the DNA. Following this repeat will be a series of six adenine nucleotides. When this region is translated, the inverted repeat (usually rich in G-C) will anneal, creating a hairpin loop structure. The six adenine nuclotides are transcribed into uracil.
A protein associated with the RNA polymerase, nusA, catches the hairpin loop structure, and holds it. This stalls transcription in the region where Uracil has been transcribed. A region of A-U in a DNA-RNA duplex (i.e., strand of RNA bound to a complementary DNA strand) is very weak (it is a major weak point). This naturally dissociates from the DNA strand, and is released by the RNA polymerase.
Summary: The intrinsic system relies on the stalling of a hairpin loop at the end of the transcript, and a region of A-U, to terminate transcription.
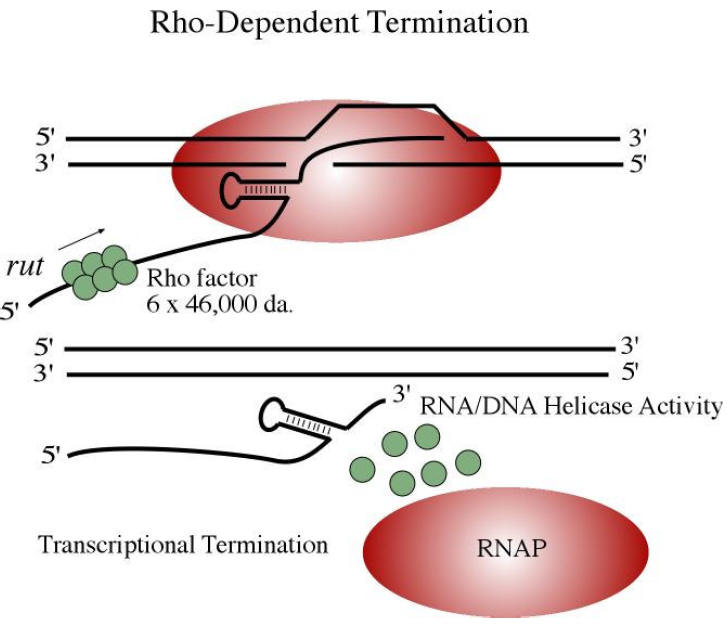
The Rho-Dependent System
The ρ-Factor is an ATP-dependent hexameric helicase, and binds to a cytosine rich region of RNA >70 bases upstream from the termination site. This cytosine dominant region is known as the Rho Utilization Site (abbreviated rut). Once bound, the ρ-Factor moves up the RNA strand toward the 3' end (e.g., where the DNA-RNA duplex is found). RNA Polymerase pauses at the termination site, and during this time, the ρ-Factor comes catches up to the transcription fork (DNA-RNA duplex), and unwinds it. Transcription ends.
Comparison between the two termination systems
 | ||
| Comparision of intrinsic termination and Rho-mediated termination. Greive S.J. and P.H. von Hippel. (2005) Thinking quantitatively about transcriptional regulation. Nat Rev Mol Cell Biol 6:221-32. |
Eukaryotic Transcription
To refresh your memory regarding Eukaryotic transcription, you may want to read over these articles:- DNA Transcription
- Transcription Factors and Transcriptional Control in Eukaryotic Cells
- Genetic Signaling: Transcription Factor Cascades and Segmentation
- Origins of New Genes and Pseudogenes
Daily Challenge
Your goal today is to discuss the differences similarities and differences between bacterial and eukaryotic transcription, specifically initiation and termination. Read DNA Transcription in case you need a refresher.REMEMBER to discuss both similarities and differences. While doing this, consider evolutionary differences.





-3-hydroxybutyrat.svg/200px-Poly-(R)-3-hydroxybutyrat.svg.png)