 Daily Newsletter
Daily Newsletter
November 12, 2013 - Lac Operon
One of the most well studied gene regulation systems is the Lac Operon found in Escherichia coli. To understand this system, it is important to first understand that bacteria do not experience Transcription and Translation in exactly the same way as eukarotes (the mechanics are very similar, but there are some distinct differences).
The core difference between prokaryotes and eukaryotes is the nucleus. Since bacteria lack a nucleus, transcription takes place in the cytosol. There is no need for a tag to get out of the nucleus, so there is no need for capping. Also, there are no introns, so no need for splicing. Basically, there is no processing of RNA to make mRNA. There is no pre-mRNA. When a gene is transcribed, the transcript is mRNA.

Since there is no membrane separating transcription from translation, you can couple these to processes. As mRNA is made, it can be translated (polyribosome).
Bacteria have a single circular molecule of DNA (a genophore, not a chromosome). They have to conserve their genetic space, so bacteria combine genes for a metabolic process in a single mRNA. IMPORTANT: bacteria can combine genes for a metabolic pathway into a sequential sequence with a single promoter. Thus, when you transcribe, you get all the genes for a given metabolic pathway.
The word OPERON describes this unique arrangement of prokaryotic genes: One promoter and one operator for a given series of metabolically linked genes.

The lac Operon holds three genes that give the cell the ability to take in and use the sugar LACTOSE. [Image Note: Bacteria have two promoter regions, -10 & -35, for a given gene or operon]
For E. coli, glucose is the preferred sugar. When glucose is present, there is no need to use lactose: these genes are not transcribed. When there is no glucose, E. coli has to use other sugars. IF lactose is present, the genes for lactose utilization will be made. Conversely, if there is no lactose, they genes remain locked down.
- Glucose Present: No transcription
- Glucose Absent: Minimal transcription
- Glucose Absent, Lactose Present: Transcription of the lac operon.
- You can block the operator of the gene. This prevents RNA polymerase from making RNA.
- You can alter the promoter (or the interaction between transcription factors and DNA) to prevent binding of the Transcription Complex (RNA polymerase).
The lac operon repressor (LACI) is a protein that is constitutively (always) expressed. This indicates that the lac operon is normally turned OFF. Notice that the gene for the regulatory protein is upstream from the lac operon. The lac repressor (LacI) binds to the major groove of the DNA at the operator. As you can see in the annotated image of the repressor, you have a DNA-binding region (active site), and a regulatory domain. The regulatory domain has the ability to bind to Lactose (the inducer).
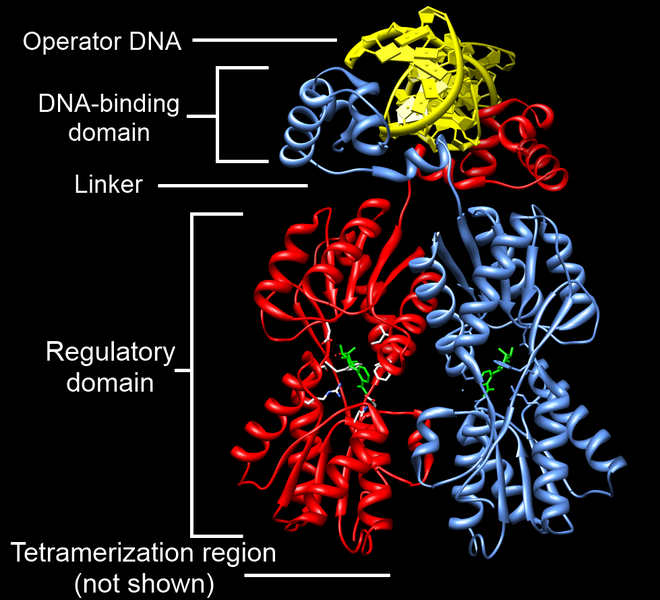 You must have a way to unlock the operon, or to put it another way, to inactivate the repressor. An inducer is a ligand that can bind to a regulatory domain, changing the shape of the regulatory domain, and thus inactivating the DNA-binding domain. In the case of the the lac repressor, the ligand is allolactose (a derivative of lactose). When allolactose binds to the lac repressor, the repressor is inactivated, and the operon cleared. This is seen in the image below.
You must have a way to unlock the operon, or to put it another way, to inactivate the repressor. An inducer is a ligand that can bind to a regulatory domain, changing the shape of the regulatory domain, and thus inactivating the DNA-binding domain. In the case of the the lac repressor, the ligand is allolactose (a derivative of lactose). When allolactose binds to the lac repressor, the repressor is inactivated, and the operon cleared. This is seen in the image below.Therefore, the lac operon is partially regulated by the presence of lactose in the environment. If there is no lactose in the environment, then there is no need to transcribe the three genes needed to use lactose.
Remember, we don't want to expend energy for things we don't need. Below is the size of the three gene products:
- β-Galactosidase 1,024 Amino Acids
- β-Galactoside Permease 418 Amino Acids
- β-galactoside transacetylase 203 Amino Acids
Tomorrow we will look at the positive regulation of the lac operon. Remember, you still will not transcribe this operon if glucose is present. You only transcribe when glucose is absent. So how does the operon know when glucose is absent? That will be our discussion tomorrow.


No comments:
Post a Comment