Initiation of Translation
Protein function is determined by the sequence of amino acids. This sequence allows the protein to fold into the correct configuration to produce activity. Any variation in the sequence can produce alterations to function, or even result in non-functionality. In order to generate the correct sequence, we must first establish the correct reading frame of codons. We must first find the start codon on mRNA.The small subunit of the Ribosome (40s in eukaryotes) is built to find the Start Codon (AUG) and will align the full ribosome with the correct reading frame. A number of proteins will help the alignment and in the formation of the full (holoenzyme) Ribosome.
The diagram below shows the overall formation of the initiation complex, complete with a tRNA (the yellow structure with a pink circle attached). Again, the function of this replication complex is to find the start codon and set the reading frame for the Ribosome. Notice that the large ribosomal subunit (60s in eukaryotes) only attaches after AUG has been found. As before, it is not necessary at this academic level to memorize all of the factors involved. What is more critical is that it is a multifactor system designed to find the correct start point, and thus the same reading frame.
Key Feature: Notice that the first tRNA is already linked to the small subunit. Why? Its anticodon is complementary to the codon on mRNA. Specifically, it has the anticodon for the start codon (AUG). So we are using base complementarity to find AUG.
Elongation
The polymerization of amino acids occurs during elongation. This is where the P and A sites become important (NOTE: P and A sites are the active sites of the enzyme). P stands for Peptidyl, while A stands for Aminoacyl. These are chemical terms, which shows the orientation of the amino acid. The exit site, represented by E, is not an active site. Consider it a disposal point for spent tRNAs. [NOTE: you may also find references to a fourth site where the tRNA first comes into the complex. Don't worry about this optional site.]The P and A sites reveal a single codon on mRNA and can hold a single complimentary tRNA. During elongation, when you have a filled P and A site, the amino acid from the P site will be linked to the amino acid in the A site. This is a process that you will have to visualize, so use the diagram below as reference:
 |
| https://karimedalla.files.wordpress.com/2012/11/translation_elongation.jpg |
The spent tRNA that started in the P site is now moved to the E site, where it is removed from the ribosome. NOTE: It takes 2 GTP to create the peptide bond, then another GTP to move the ribosome. So a total of 3 GTP are used in one 'round' of Ribosomal action. REMEMBER THIS! In addition, it took a triphosphate to charge tRNA (so a total of 4 for each amino acid added to the polymer).
When both the P and A sites have charged tRNA, the growing chain from the P site is added to the single amino acid in the A site. The ribosome shifts and the process continues. This elongation process of adding amino acids (amino acid polymerization) will continue until a STOP codon is reached (UAA, UAG, and UGA).
Question: How much ATP will you need to expend to make a protein with 100 amino acids? How about a 150 amino acid protein? GOAL: Recognize and be able to articulate why protein synthesis is an energy consumptive process, and be able to discuss why it is critical for cells to regulate energy consumptive processes.
Termination
To create a functional protein, translation must end with the appropriate amino acid. If translation stops to soon, the protein will be too short and may not bend (configure) correctly. If it is too long, then it may not bend (configure) correctly. Termination is a critical process. Termination begins when a STOP CODON (UAA, UAG, and UGA) is reached. In eukaryotes, a releasing factor is used to separate the ribosomal subunits. KEY CONCEPT: The stop codon signals the end of the coded message. |
| http://www.proteinsynthesis.org/wp-content/uploads/2013/06/protein-synthesis-steps-termination.jpg |
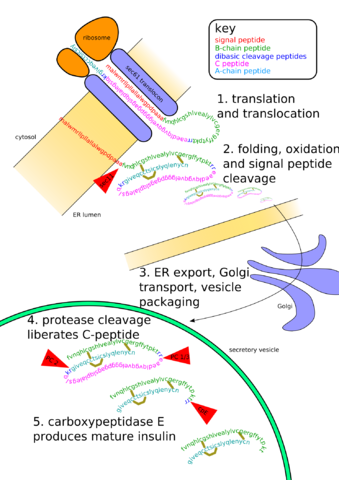
Once completed, proteins can be further modified as fits their function (such as adding sugars). This is known as post-translational modification. The image below shows the posttranslational modifications needed in the production of insulin. Production starts with a ribosome-bound on the Rough Endoplasmic Reticulum (RER). Processing will occur in the RER and in the Golgi body. This is only one example of posttranscriptional modification, and a majority of proteins require such modifications before they are functional. [NOTE: as a general rule, there is a less extensive posttranscriptional modification in prokaryotes, but they have numerous proteins that do require modification].
Daily Challenge
In your own words, describe the process of translation. Discuss initiation, elongation, and termination. Make sure that you discuss the P and A site, as well as the importance of the start and stop codon. Afterwards, give a BRIEF discussion on how this is an energy consumptive process that needs to be regulated.Optional Challenge: Genomics and Proteomics
Read the following articles:A brief guide to genomics - NIH fact sheet
Transcriptome - NIH fact sheet
Proteomics
The genome can be seen as the genetic potential of an individual (think Genotype), while the proteome shows what is actually produced at a given time, under a given condition (consider this the phenotype). Provide a discussion of the importance of genomic and proteomic studies in modern biological research, and make sure that you provide a description of both the genome and the proteome of an organism.
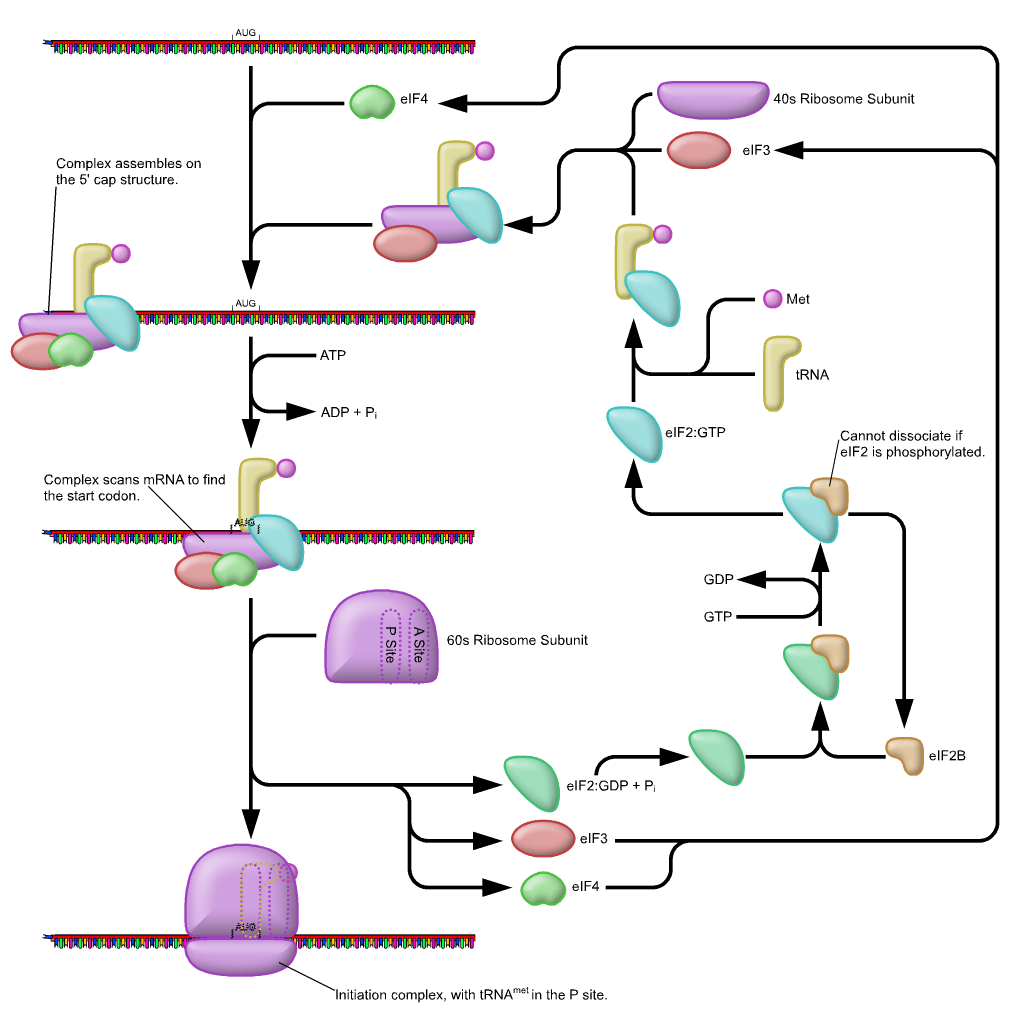
This blog post will explain the difference between PoC, prototype, and MVP and how to test the assumptions with the minimum set of features. We'll go through each approach in detail and show how the process looks like in practice. https://cxdojo.com/poc-prototype-or-mvp-how-to-achieve-product-market-fit
ReplyDelete